Arnold Wiliem, YongKang Wong, Conrad Sanderson, Peter Hobson, Shaokang Chen, Brian C. Lovell
Sullivan Nicolaides Pathology, Australia
School of ITEE, The University of Queensland, Australia
NICTA, Australia
[2013-09-23] Cell images are now available!
[2013-09-12] Patch-level features are now available!
SNPHEp-2 Dataset [description and files]
Features + Codes [description and files]
The Anti-Nuclear Antibody (ANA) clinical pathology test is commonly used to identify the existence of various diseases. A hallmark method for identifying the presence of ANAs is the Indirect Immunofluorescence method on Human Epithelial HEp-2 cells, due to its high sensitivity and the large range of antigens that can be detected. However, the method suffers from numerous shortcomings, such as being subjective as well as time and labour intensive. Computer Aided Diagnostic (CAD) systems have been developed to address these problems, which automatically classify a HEp-2 cell image into one of its known patterns (eg. speckled, homogeneous). Most of the existing CAD systems use handpicked features to represent a HEp-2 cell image, which may only work in limited scenarios. In this paper, we propose a cell classification system comprised of a dual-region codebook-based descriptor, combined with the Nearest Convex Hull Classifier. We evaluate the performance of several variants of the descriptor on two publicly available datasets: ICPR HEp-2 cell classification contest dataset and the new SNPHEp-2 dataset. To our knowledge, this is the first time codebook-based descriptors are applied and studied in this domain. Experiments show that the proposed system has consistent high performance and is more robust than two recent CAD systems.
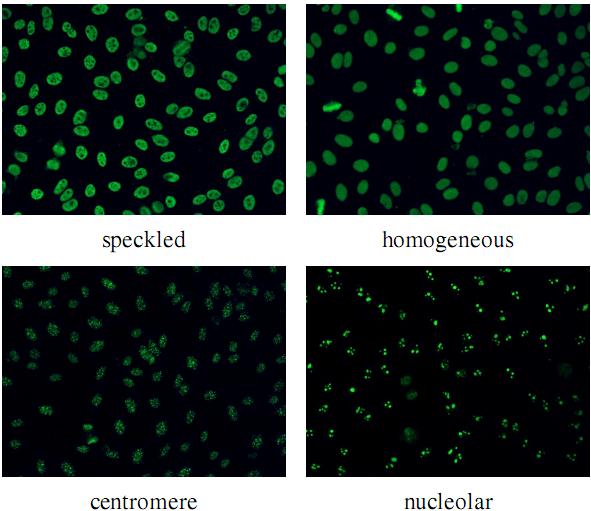
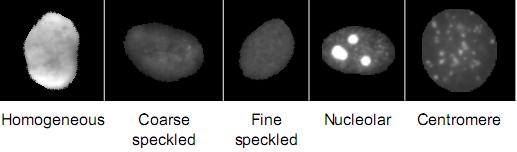
The SNP HEp-2 Cell Dataset (SNPHEp-2) was obtained between January and February 2012 at Sullivan Nicolaides Pathology laboratory, Australia. The dataset has five patterns: centromere, coarse speckled, fine speckled, homogeneous and nucleolar. The 18-well slide of HEP-2000 IIF assay from Immuno Concepts N.A. Ltd. with screening dilution 1:80 was used to prepare 40 specimens (see figure belows). Each specimen image was captured using a monochrome high dynamic range cooled microscopy camera, which was fitted on a microscope with a plan-Apochromat 20x/0.8 objective lenses and an LED illumination source. DAPI image channel was used to automatically extract the cell image masks.

There are 1,884 cell images (see figure belows) extracted from 40 specimen images. The specimen images are divided into training and testing sets with 20 images each (4 images for each pattern). In total there are 905 and 979 cell images extracted for training and testing. Five-fold validation of training and testing were created by randomly selecting the training and test images. Both training and testing in each fold contain around 900 cell images (approx. 450 images each).
The SNPHEp-2 dataset and features + codes ('Licensed Material') are made available to the scientific community for non-commercial research purposes such as academic research, teaching, scientific publications or personal experimentation. Permission is granted by National ICT Australia Limited (NICTA) to you (the 'Licensee') to use, copy and distribute the Licensed Material in accordance with the following terms and conditions:
A.Wiliem, Y. Wong, C. Sanderson, P. Hobson and B.C. Lovell
IEEE Workshop on Applications of Computer Vision, WACV 2013
Bibtex entry:
@INPROCEEDINGS{wiliem_wacv_2013,
AUTHOR = {Arnold Wiliem and Yongkang Wong and Conrad Sanderson and Peter Hobson and Shaokang Chen and Brian C. Lovell},
TITLE = {Classification of Human Epithelial Type 2 Cell Indirect Immunofluoresence Images via Codebook Based Descriptors},
BOOKTITLE = {IEEE Workshop on Applications of Computer Vision (WACV)},
PAGE = {95--102},
YEAR = {2013}
}
Cell images & Masks [download] (3 Megabytes)
Patch level features [download] (1 Gigabytes)
Histogram level features [coming soon...]
Convex hull classifier [coming soon...]
Further enquiries can be made to {a döt wiliem ät uq döt edu döt au} or {lovell ät itee döt uq döt edu döt au}